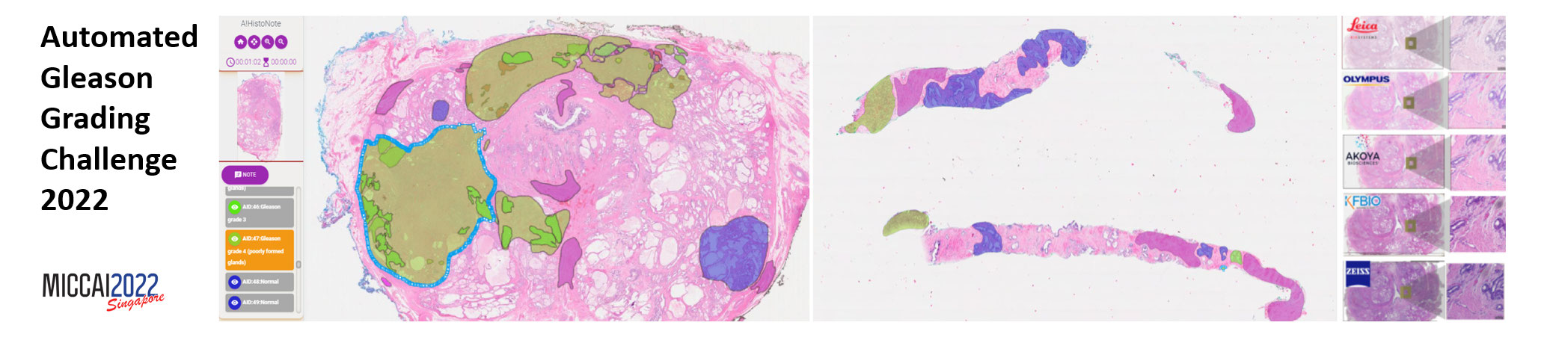
Automated Gleason Grading Challenge 2022¶
Important updates¶
**10/5/2024 :We are excited to share that our paper, based on the dataset, has now been published. We kindly request that any use of this dataset be accompanied by a citation of our paper. Thank you! **¶
18/5/2023 :** **The preprint of our paper (our own study on the dataset, NOT the challenge paper) is available here.
25/10/2022 ! Announcement! :** **As a part of MICCAI2022 challenge, the registration of our change was closed in May and the final rankings was announced in August. Although the MICCAI2022 challenge has ended, we will publish the complete data set in order to share our data with researchers in the field. Please fill the form and the download links will be sent to you within 15 working days.
Additionally, we will make the challenge alive by adopting "Post Challenge Phase". Registered participants (the registration window has been re-opened) can submit their results via the submission window to quickly validate their results. Alternatively, participants are encouraged to design other evaluation metrics. Please note that there will be no award for Post Challenge Phase.
Last but not least, we are planning to collect some very interesting data to quantitatively study the impact of annotation qualities. Users who sign up for the complete dataset will receive the update via email. Please check our website once a while if you're interested : )
12/10/2022 - The complete data set will be published soon.
4/10/2022 - Post Challenge Phase is open. Participants can submit their results for evaluation.
22/9/2022 - We are planning to publish the complete dataset. Meanwhile, if you are interested to the challenge data, please fill the form (12/10/2022 - The complete data set will be published soon. This form has been closed) and a download link will be sent to you within 5 working days. Thank you : )
Motivation¶
Prostate cancer is one of the most common cancers in men around the world. The Gleason grading system remains the most powerful prognostic predictor for patients with prostate cancer since the 1960s. The method is to evaluate lesion levels of Prostate cancer via the microscopic inspection of stained biopsy tissue and estimate the Gleason score. However, it is time-consuming for pathologists to identify the cellular and glandular patterns for Gleason grading. Moreover, its application requires highly-trained pathologists, is tedious, and yet suffers from limited inter-pathologist reproducibility. With the development of digital scanners, the computer-aided diagnosis method based on a convolutional neural network has played an important role in medical image segmentation and detection. To help drive forward research and innovation for Automated Gleason Grading in computational pathology, we organize the Automated Gleason Grading Challenge (AGGC) 2022. The challenge requires researchers to develop algorithms that identify distinct Gleason Patterns within the H&E-stained whole slide image dataset.
AGGC2022 has been accepted into MICCAI 2022 challenges (http://www.miccai.org/special-interest-groups/challenges/miccai-registered-challenges/). This challenge will showcase the best automated Gleason grading techniques that will work on a diverse set of H&E stained histology images obtained from different digital scanners. To the best of our knowledge, this is the first challenge in the field of digital pathology that investigate the variations caused by image scanning.
Assessment aim¶
Find automated Gleason grading algorithm with high accuracy for H&E-stained prostate histopathological images scanned by multiple scanners.
Challenge Keywords: Prostate Cancer; Gleason Patterns; Digital Pathology; Deep Learning¶
Participation¶
Please click the "Join" button on the top right corner of this page. If you do not have an account at grand-challenge.org, please sign up. If you are participating in a team of more than two members, please use only one ID to register for the contest.
Important dates¶
The release date of the training data: Apr 1, 2022
The registration period: Apr 1 – May 25, 2021
The release date of the public test data: Jun 15, 2022
The submission period: Jun 30 – Aug 10, 2022
The official release date of the results: Aug 26, 2022
Prizes (sponsored by AWS and HUAWEI)¶
1st place: AWS Cloud Credit worth \$2,500 + HUAWEI Cloud Credit worth \$1,500
2nd place: AWS Cloud Credit worth \$1,500 + HUAWEI Cloud Credit worth \$750
3rd place: AWS Cloud Credit worth \$1,000 + HUAWEI Cloud Credit worth \$750
4th place: HUAWEI Cloud Credit worth \$500
5th place: HUAWEI Cloud Credit worth \$500
6th place: HUAWEI Cloud Credit worth \$500
The top 3 performing methods will be announced publicly.
Citation¶
If you use our dataset, please cite our paper (under preparation). The preprint is available here.
Publication¶
We will publish a paper to describe the challenge as well as the methods and results of the top 5 participating teams. 2-3 members from the selected teams will be invited to contribute to the manuscript and qualified as authors. The participating teams may publish their own results separately after the challenge paper is published.
Sponsors¶
This challenge is sponsored by AWS and HUAWEI.
Click here to learn more about** Medical Imaging Solutions with AWS**.
Click here to learn more about HUAWEI CLOUD.